paired end sequencing read length
The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. Enter up to 20 characters or use manual mode if you need between 20 and 100 bp.

How To Calculate The Coverage For A Ngs Experiment
When using RNA-Seq to study gene expression read length is not a significant factor.

. One from either end see figure 1a. What matters is read counts. When the fragment length is longer than the combined read length.
Read Introduction Paired-End DNA Sequencing Paired-end DNA sequencing reads provide superior alignment across DNA regions containing repetitive sequences and produce longer. During paired-end sequencing two sequencing reads are generated for each library molecule. The maximum distance x for a pair considered to be properly paired SAM flag 02 is calculated by solving equation Phi x-musigmaxLp0 where mu is the mean.
In paired-end reading it starts at one read finishes this direction at. The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. Application of sequencing to RNA analysis RNA-Seq whole transcriptome SAGE expression analysis novel organism mining splice variants Search in titles only Search in RNA.
In single-end reading the sequencer reads a fragment from only one end to the other generating the sequence of base pairs. If the read length is chosen significantly larger than half the. The library prep protocols are designed to.
Reading the nucleotides from two ends of a DNA fragment is called paired-end tag PET sequencing. We use an Illumina MiniSeq for our short-read sequencing runs. The paired-end short read lengths are always 2 x 150bp 300bp.
Whether you align 100bp paired reads or a. To ensure sequencing quality of the Index Read do not exceed the supported read.
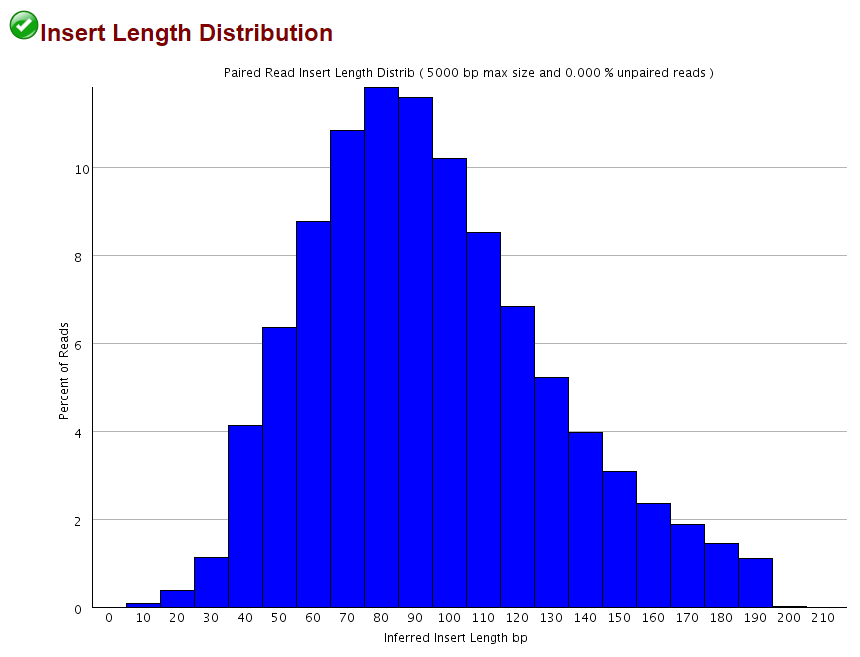
Qc Fail Sequencing Mispriming In Pbat Libraries Causes Methylation Bias And Poor Mapping Efficiencies
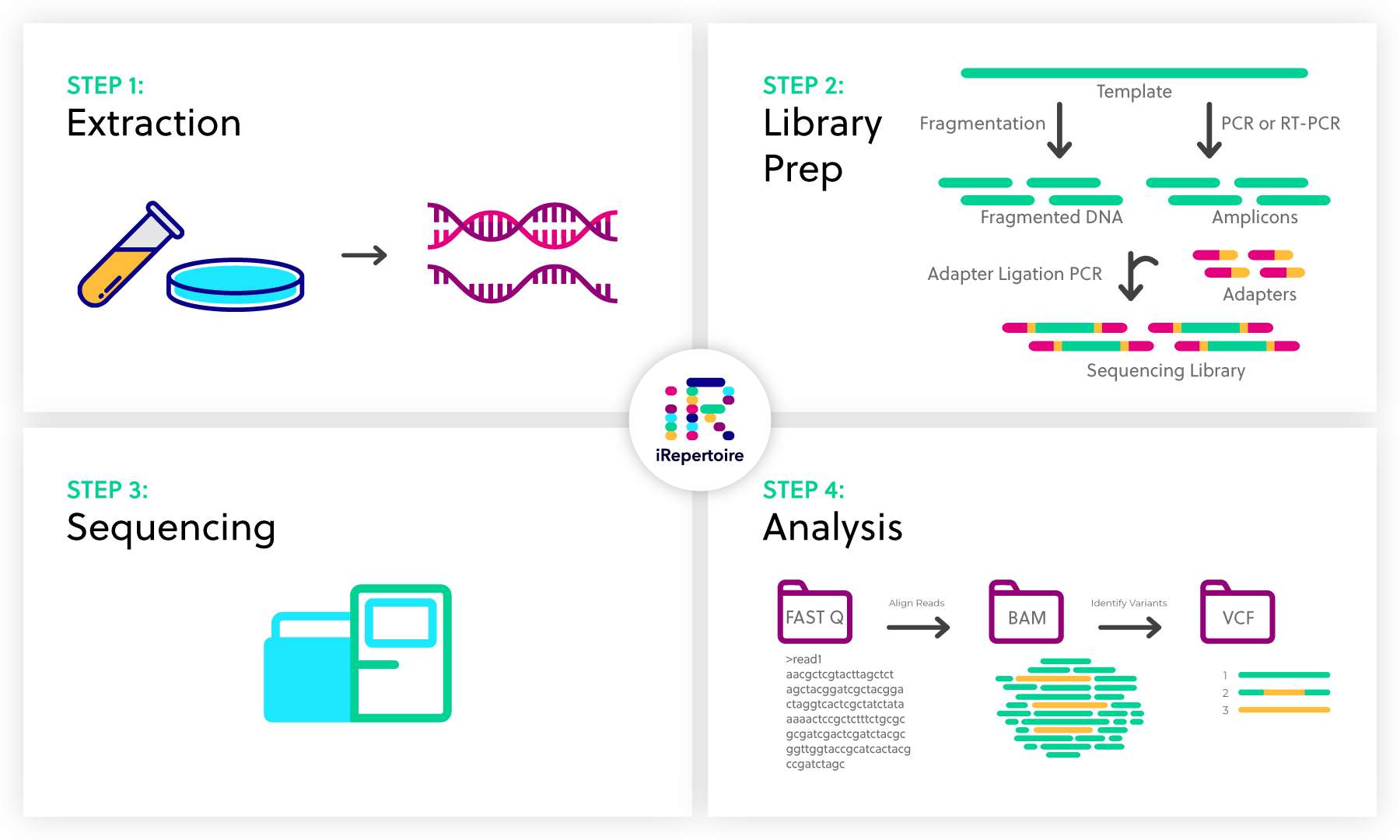
Next Generation Sequencing Ngs Overview Irepertoire Inc

Sequencing Read Length How To Calculate Ngs Read Length
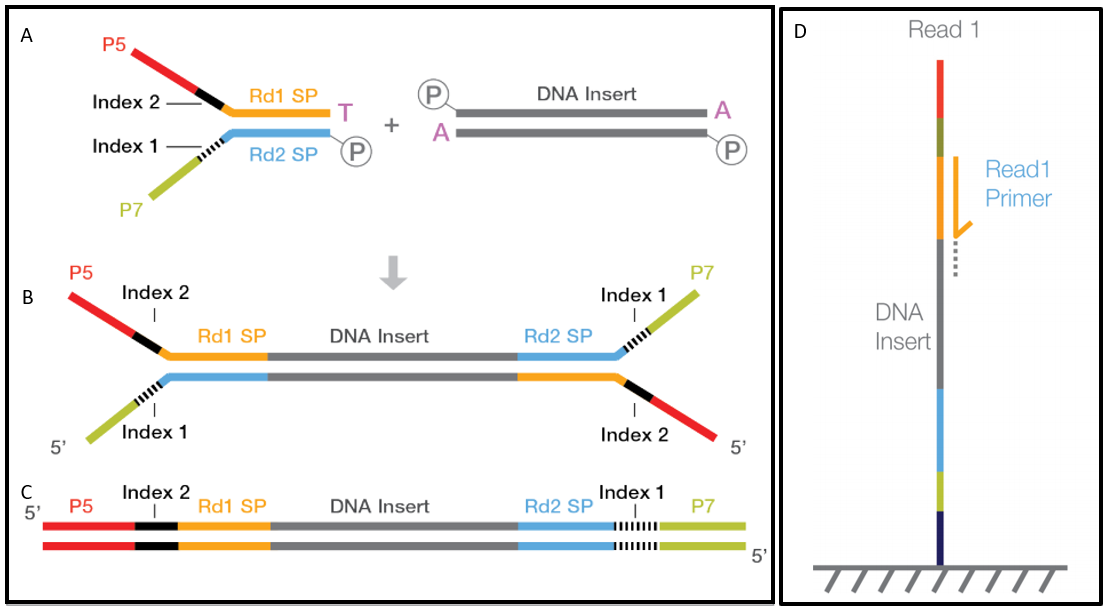
How Short Inserts Affect Sequencing Performance

What Are Paired End Reads The Sequencing Center
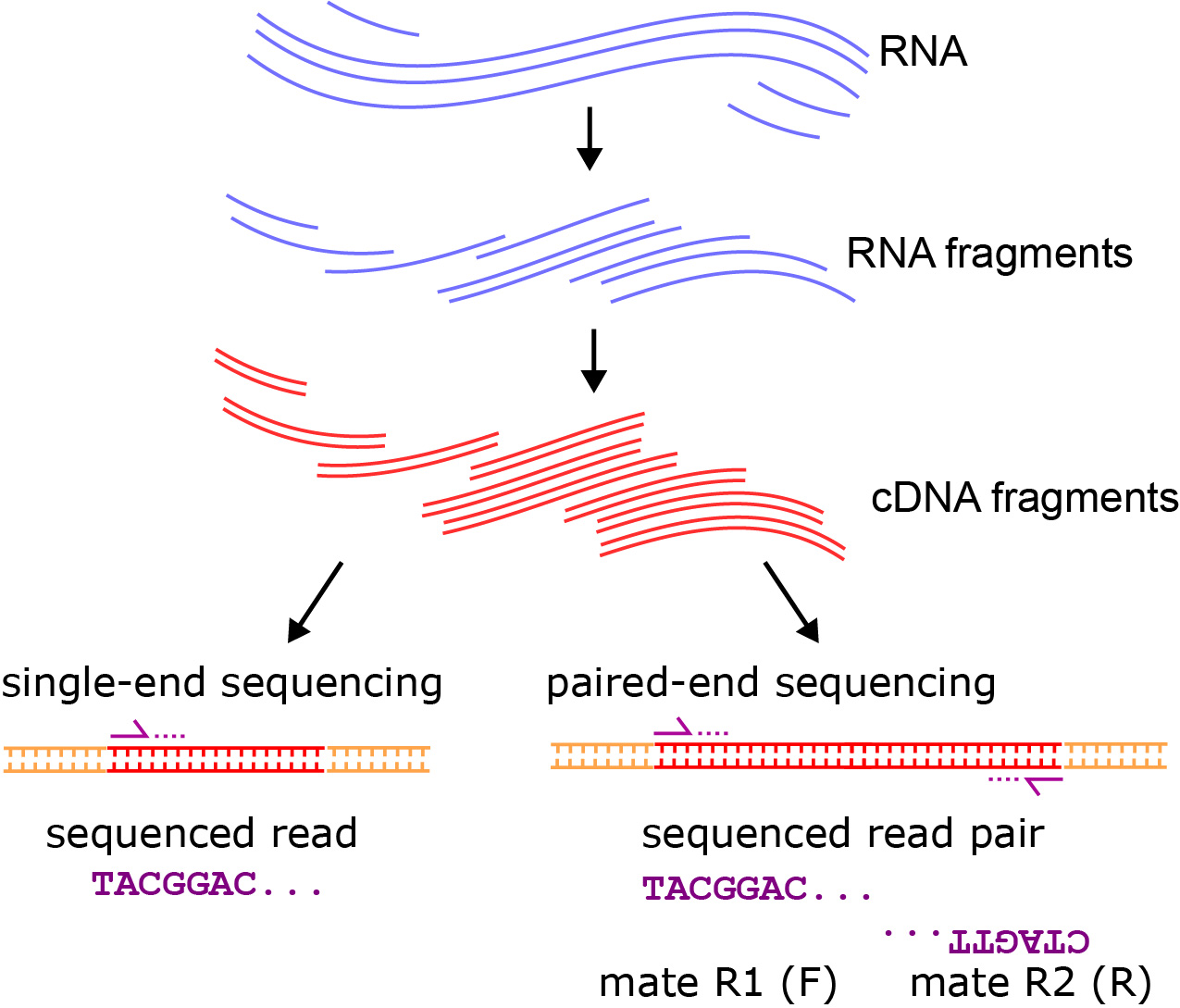
Chapter 6 Transcriptomics Applied Bioinformatics

Rapid Low Cost Assembly Of The Drosophila Melanogaster Reference Genome Using Low Coverage Long Read Sequencing Biorxiv
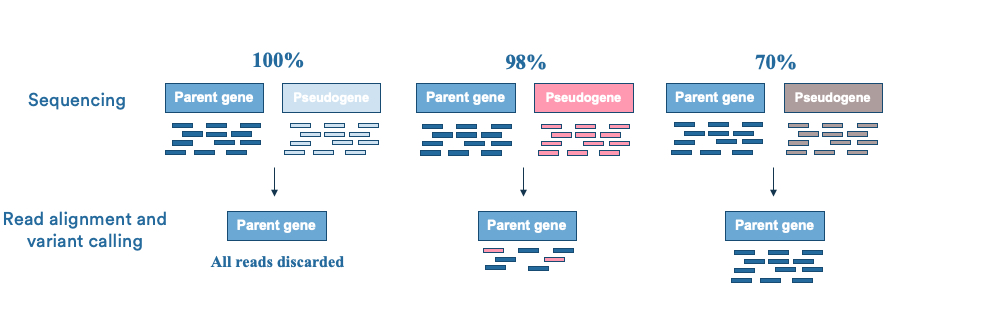
Blueprint Genetics Approach To Pseudogenes And Other Duplicated Genomic Regions Blueprint Genetics
Single End Paired End Mate Pair 네이버 블로그

What Is Mate Pair Sequencing For

Dnbseq G50 Gets Full Performance Upgrade Mgi Leading Life Science Innovation

Next Generation Sequencing In Aquatic Models Intechopen
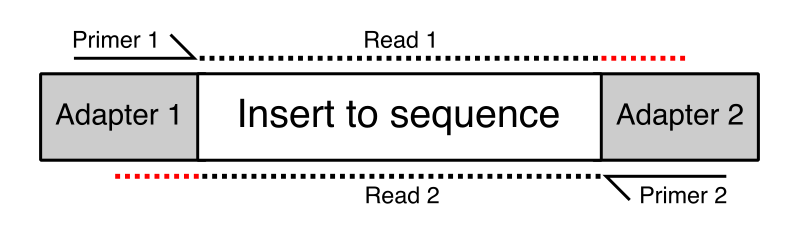
Qc Fail Sequencing Read Through Adapters Can Appear At The Ends Of Sequencing Reads

Next Generation Sequencing Tips N Tricks Part 4 Diagnostech
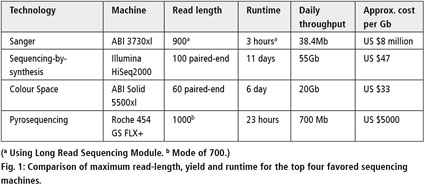
Analyzing Next Generation Sequence Data 2011 Wiley Analytical Science


